Next-Generation CRISPR-Based T-Cell Therapies: Opportunities and Challenges
T cells can be reprogrammed to selectively recognize and eliminate cancer cells, and such T-cell therapies have shown substantial efficacy in patients with specific hematologic malignancies. Nevertheless, significant challenges persist, including primary and secondary resistance, limited effectiveness against solid tumors, a narrow spectrum of targetable antigens, and complex, time-consuming manufacturing processes.
CRISPR-based genome editing offers new avenues to enhance the antitumor potency of T cells. Base and prime editing, often collectively referred to as CRISPR 2.0, provide greater precision and efficiency than conventional CRISPR methods. Rather than introducing double-strand DNA breaks, these technologies enable the targeted and highly accurate rewriting of gene sequences.
In our review published in Nature Reviews Clinical Oncology, conducted in collaboration with colleagues from multiple research institutions, we explore both the challenges and the promising opportunities associated with implementing CRISPR 2.0 technologies in T-cell therapy. We discuss emerging CRISPR-2.0 approaches, recent progress toward clinical translation, and strategies for harnessing these technologies to further advance cellular immunotherapies.
Karl Petri, Elvira D’Ippolito, Annette Künkele, Ulrike Köhl, Dirk H. Busch, Hermann Einsele & Michael Hudecek. Next-generation T cell immunotherapies engineered with CRISPR base and prime editing: challenges and opportunities. Nat Rev Clin Oncol (2025). https://doi.org/10.1038/s41571-025-01072-4
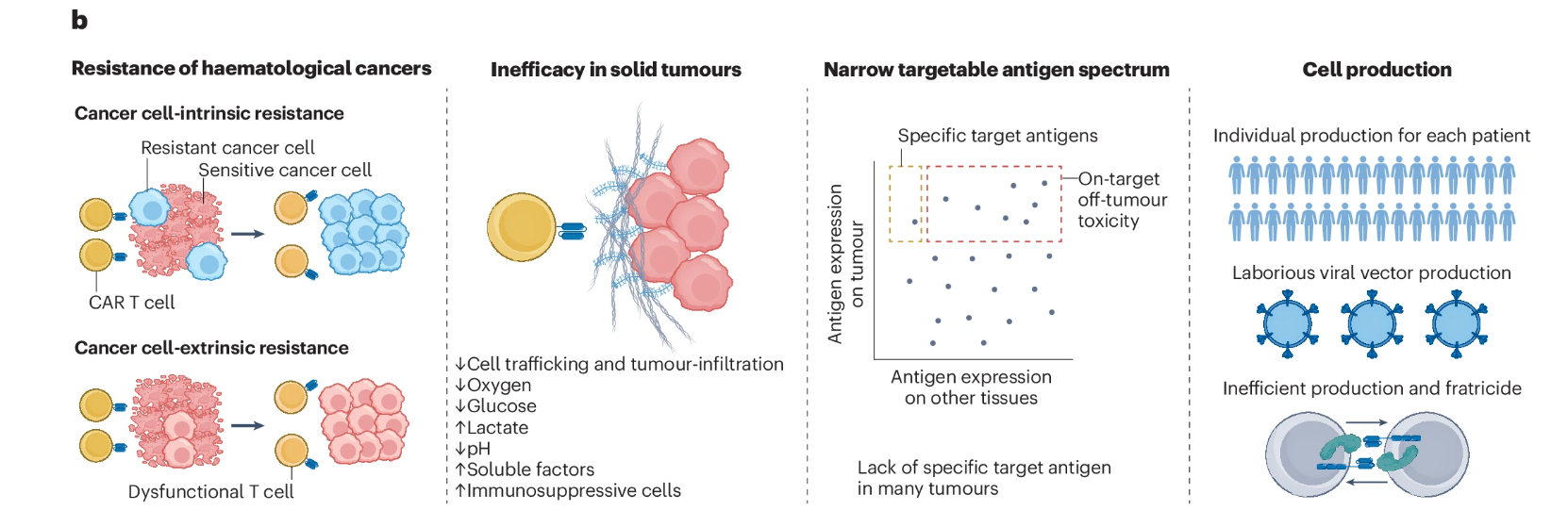
Schematic overview of the major challenges in cellular immunotherapy in oncology and how each could potentially be addressed using novel CRISPR 2.0–based gene-editing strategies. BCMA, B-cell maturation antigen; PE, prime editing; TALEN, transcription activator–like effector nuclease; TCR, T-cell receptor; ZFN, zinc-finger nuclease.
